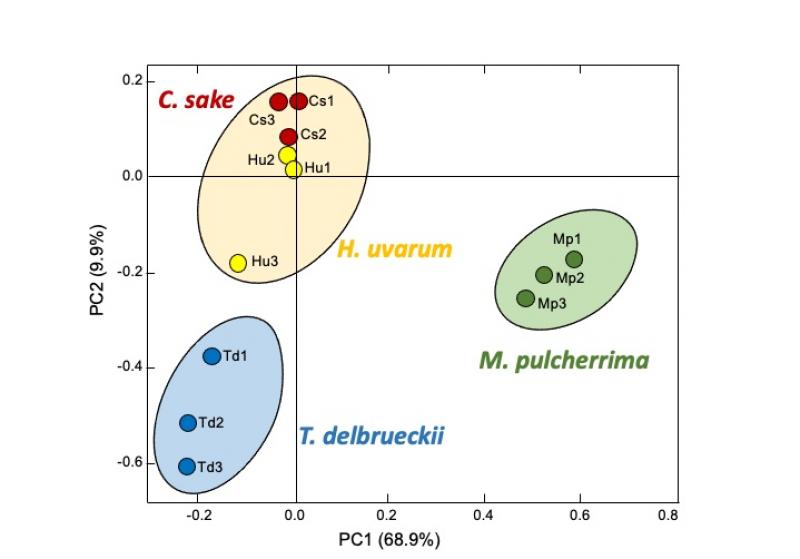
One of the main trends in oenological biotechnology in the last decade has been the incorporation of a growing number of non-Saccharomyces strains (alternative species to Saccharomyces cerevisiae) into oenological yeast catalogs. These yeasts provide several advantages, especially for their contribution to the aromatic composition of the wines. In most cases they are used in combination with S. cerevisiae to ensure an adequate end of fermentation. This is why the ICVV's Microwine group has been studying for years the interactions between various oenological yeasts and the biological mechanisms involved. In previous works we observed, an acceleration of glycolysis by S. cerevisiae, among other effects, when confronted with yeasts such as Torulaspora delbrueckii, Candida sake or Hanseniaspora uvarum. These changes affect the transcription of certain genes and were observed in very short response times (3 hours).
In our last work, it was observed that the response of S. cerevisiae to another common yeast in wine, Metschnikowia pulcherrima, besides sharing similarities with the previous ones, has the particularity of causing a reduction in the transcription of genes related to respiration. Our work suggests that there is a specific recognition among yeasts, and a more or less standardized response by S. cerevisiae (that we have called "direct interaction stress response”) but modulated according to the specific strain it faces. This might be part of the set of mechanisms that S. cerevisiae has to displace other yeasts during fermentation. A better understanding of these interaction mechanisms will help us to develop new yeast combinations for better winemaking control, especially when using non-Saccharomyces yeasts.
This study was recently published in the journal Food Microbiology. More information:
https://www.sciencedirect.com/science/article/pii/S0740002020302598
Free access to the full text for a limited time by following this link: